miRNA-Seq Analysis Workflow
In this tutorial, we will introduce the miRNA-Seq data analysis workflow in ArrayStudio, step by step. The workflow consists of a number of modules for miRNA-Seq data processing, including raw data quality control (QC), adapter trimming, alignment, aligned data QC, quantification, and differential expression
This workflow can be followed by the steps listed in the Workflow window:
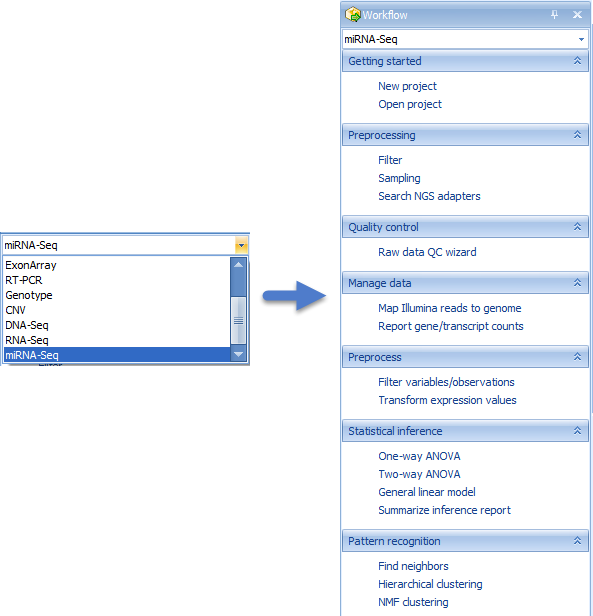
or by running the individual modules from the menu items.
For convenience, especially for new users, we also have a single-command pipeline that will use default settings to take miRNA-seq data from raw fastq files to mapped, QC'd, and quantified data, which follows the workflow below:
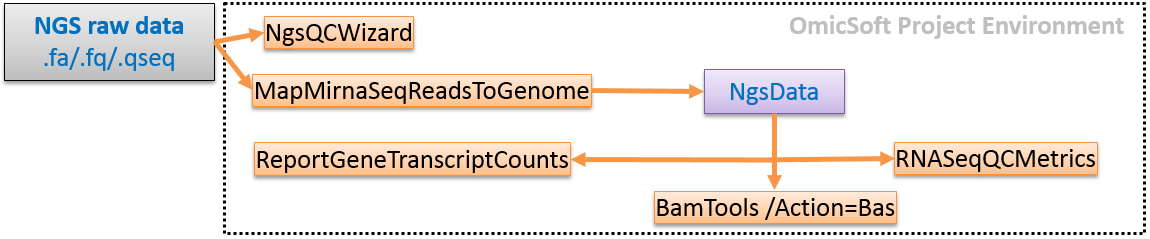
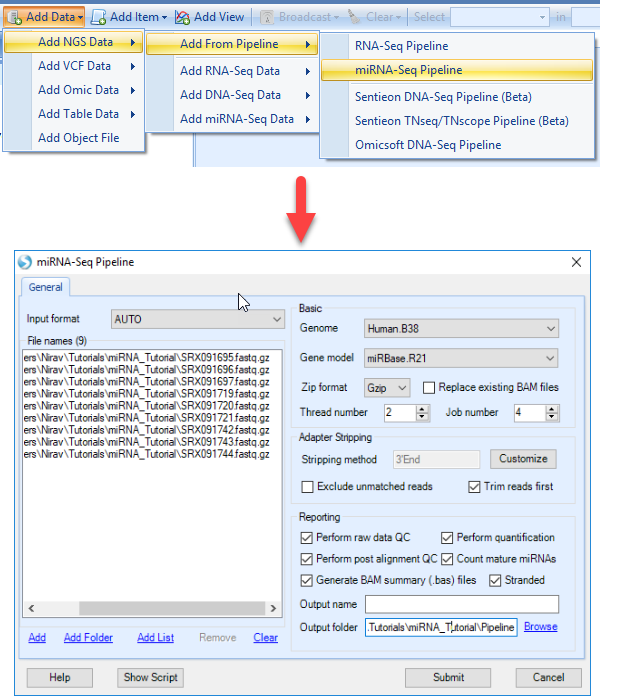
In this tutorial, we will go step by step through all the options specified in this pipeline to give users a detailed understanding of each option selected above and how they can impact your study.