Mutation Filters
For views that contain data from DNA-seq and RNA-seq, these views can be filtered to variants of interest. The Mutation tab will appear in these views for filtering:
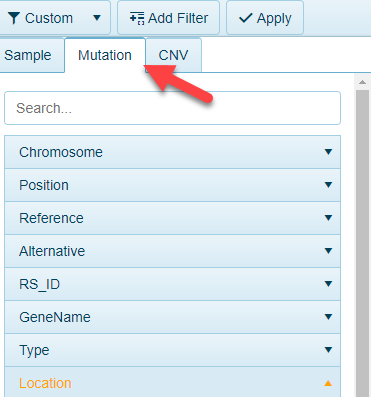
The filters can be applied to:
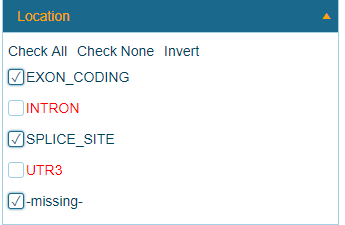
Location
By default, only non-synonymous coding mutations are applied in the Land. Users can filter to intronic and 3'UTR variant calls:
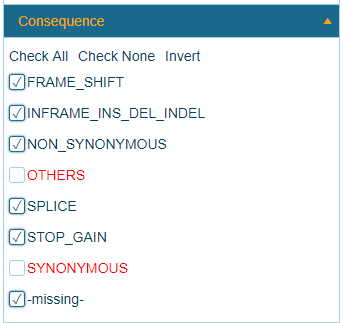
Consequence
By default, only non-synonymous coding mutations are applied in the Land. Users can filter synonymous mutations, or perhaps more stringent filters using this mutation metadata:
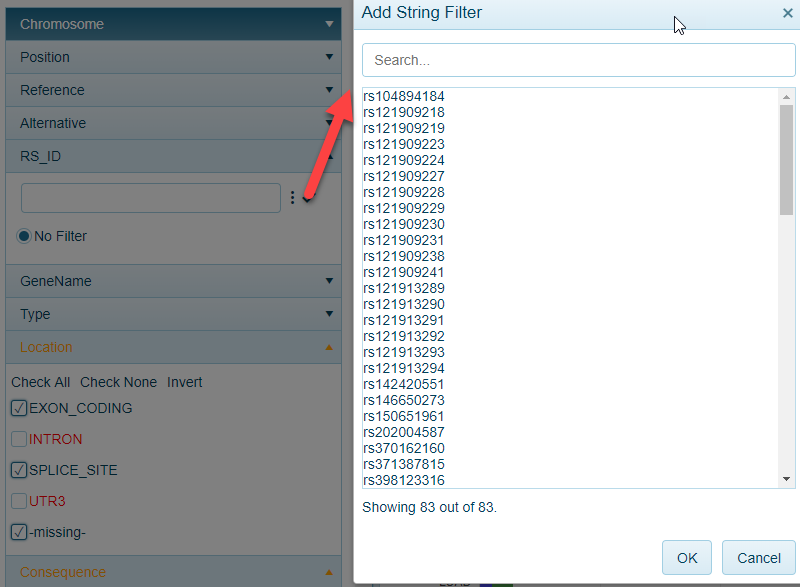
RS_ID
RS_ID (when available) can be used to filter to specific mutations:
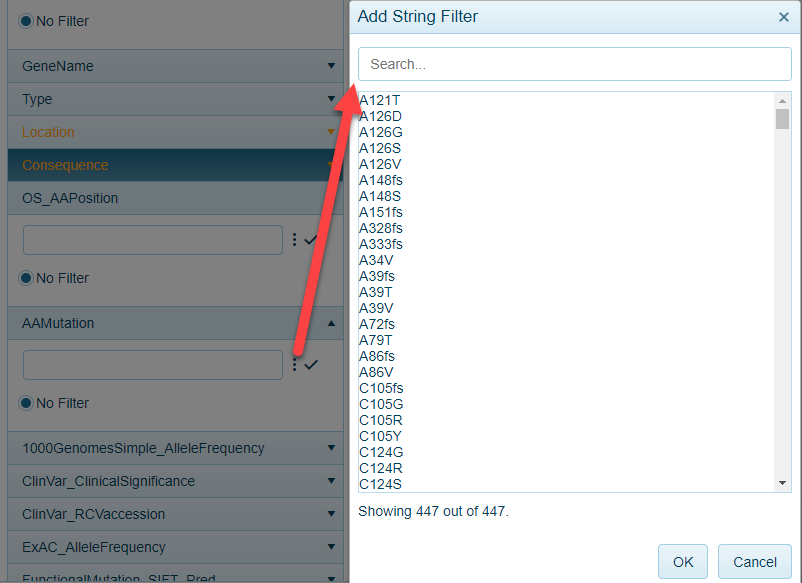
AAMutation (Amino Acid Mutation)
All variants in a searched gene(s) are represented in the mutation views. Users can filter to specific variants using this filter:
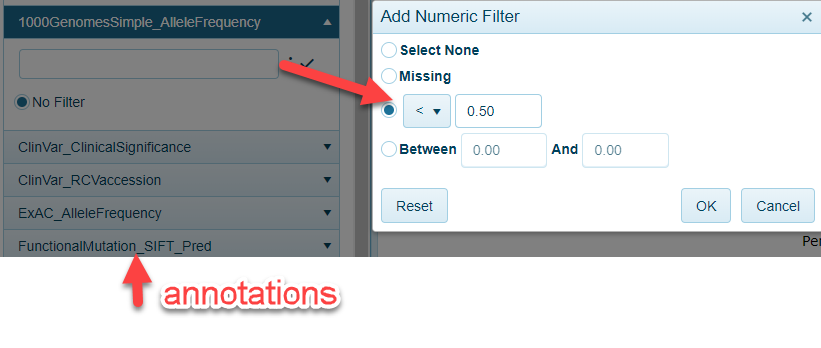
Classifiers
A number of classifiers can be used to filter for variants that are not highly abundant in variant databases. For example, in the case below, users can filter to mutations in a gene with less than fifty percent frequency in 1000 Genomes: