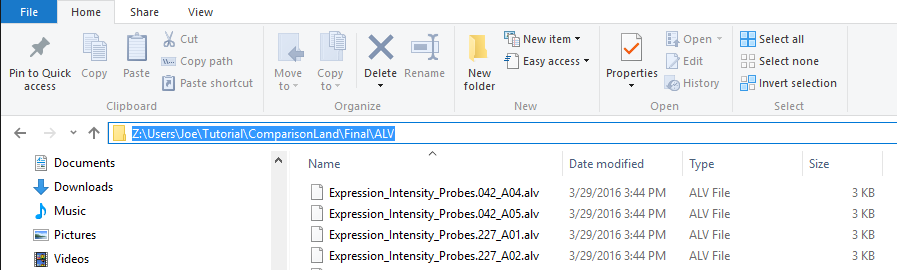
Convert Expression Data to ALV
The first step in creating your own ComparisonLand is to import your data into Array Studio and attach Annotation and Design metadata. The resulting .osobj file will be converted to Land-compatible .alv files.
Create Array Studio Project
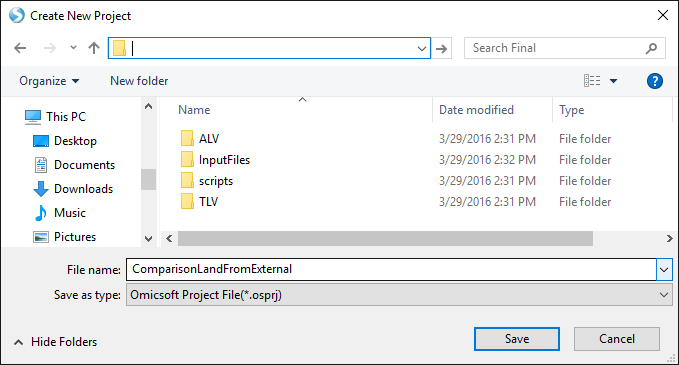
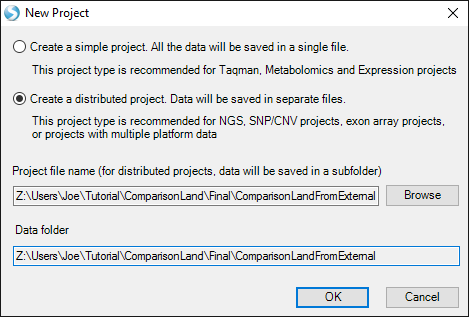
First, open Array Studio, and create a new distributed project:
Import Expression Data into Array Studio
The tutorial data are expression intensity measurements from an Affymetrix array for ~100 probesets (genes). Each row contains all of the measurements for one probeset, with each sample in columns.
To import these data, click Add Data | Expression Data:
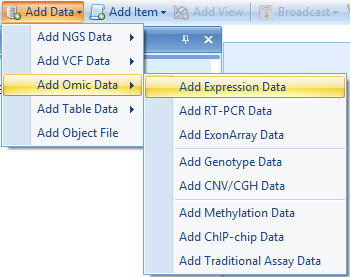
The tutorial data are tab-delimited, one gene per row, but Array Studio can import data from a variety of sources:
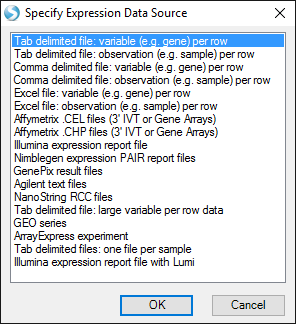
If your data have additional annotation columns (besides the ProbeSetID in the first column), you can specify them, and they will be automatically added as metadata. The tutorial data do not have any extra columns, so do not select anything, and press OK:
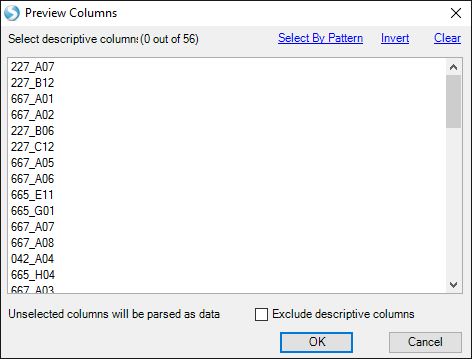
You will be prompted to add a Design metadata file, but you may add one later if you prefer:
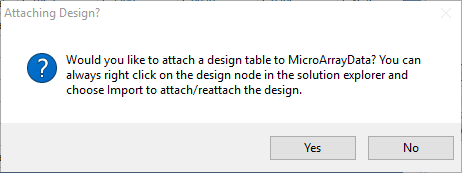
For this tutorial, select Yes, then select Tab delimited File, navigate to "Brawndocin_Slurmycin.V2.Expression.Design.txt", and click OK without appending to the existing covariate table.
You will now be prompted to add Annotation metadata. Again, you may add one later, but for the tutorial dataset you can add it immediately:
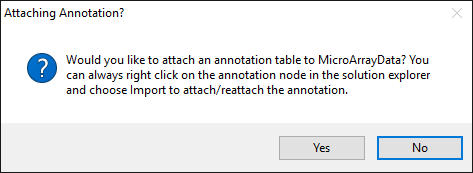
The tutorial dataset probesets are from Affymetrix HG-U133A, which is one of many online annotations available from Array Studio.
Select source Online annotation file
and search for HG-U133A, which will list all matching annotations:
Select OK, which will automatically retrieve the annotation and attach it to the data. Click OK on the subsequent window without selecting either option.
Now you will see the tutorial data imported as an -Omic data type, which is a tabular dataset with associated Design and Annotation data:
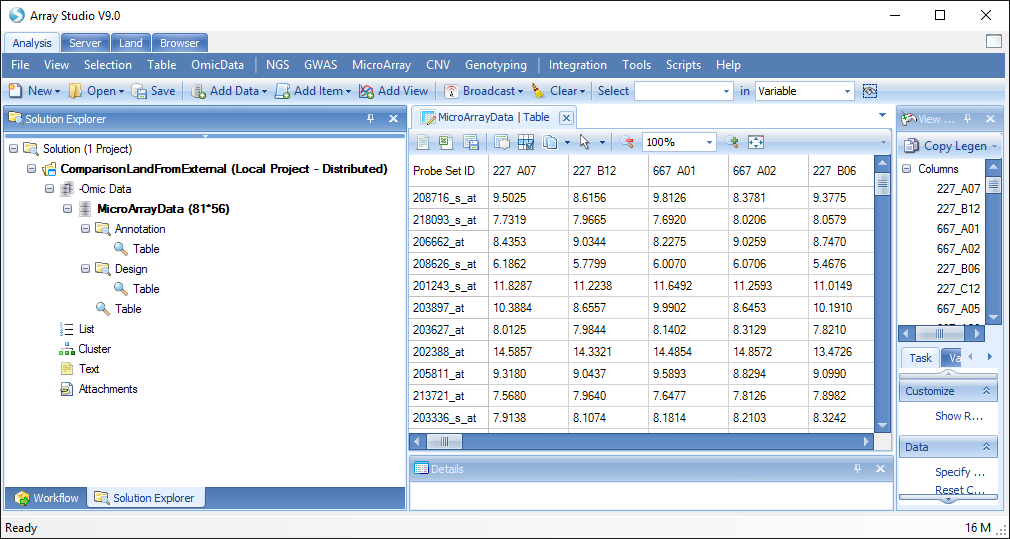
In the Solution Explorer, right-click on MicroArrayData and select Rename, then rename the -Omic object to "Brawndocin_Slurmycin.Expression":
Your Solution Explorer will update the object name:
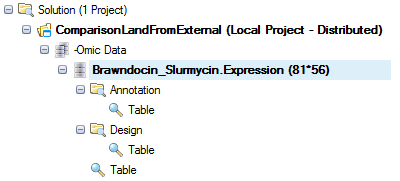
Now save the project. The .osobj file "Brawndocin_Slurmycin.Expression.osobj" will be used to create ComparisonLand .alv files.
Convert Expression .OSOBJ file to Land-compatible .ALV files
Now you can convert the .osobj file to .alv files, which can be added to your ComparisonLand.
To do this, you will run an OmicScript (Oscript) from Array Studio.
Oscripts are text-format files that instruct Array Studio to perform commands. All Array Studio GUI functions have associated .Oscript functions. There are additional "Oscript-only" functions (such as ConvertExpressionOsobj), which do not have a GUI module window in Array Studio.
An example Oscript is ExtractExpressionOsobjToAlv.oscript, which you will find in the zipped tutorial set: ::
Begin LandTools /Namespace=NgsLib;
Files "Z:\Users\Joe\Tutorial\ComparisonLand\Final\
ComparisonLandFromExternal\
Brawndocin_Slurmycin.Expression.osobj";
Reference Human.B37.3;
GeneModel OmicsoftGene20130723;
Options /Action=ConvertExpressionOsobj
/SampleIDColumn="Observation"
/MappingID=Affymetrix.HG-U133A_Human.B37.3
/IsRatio=False /MedianNormalization=False
/TargetMedian=0
/OutputFolder=
"Z:\Users\Joe\Tutorial\ComparisonLand\Final\ALV";
End;
For the tutorial, will need to change the input and output paths to match where your .osobj file is located, and where you want to output the .ALV files.
Once you have modified and saved the Oscript file, Select Tools | Run Script:
Navigate to your modified ExtractExpressionOsobjToAlv.oshell.oscript file, and click OK.
The output .ALV files will be found in the folder you specified. You will find one file per sample: